Lecture 2: Command Line Basics¶
CBIO (CSCI) 4835/6835: Introduction to Computational Biology
Overview and Objectives¶
In this lecture, we'll eschew all things Python and Biology, and focus entirely on the step before either of these: becoming familiar with the command line (or command prompt). By the end of this lecture, you should be able to:
- Name the different kinds of command line "shells"
- Navigate through the folders of a filesystem
- Perform basic text parsing using bash commands
Part 1: BASH Basics¶
If you've never used a command-line before... Don't be intimidated!
Bash is to command prompts as Windows is to operating systems¶
Other command prompts include
csh(some would say the original: the "C-shell"bash("bourne-again" shell; tends to be default on most Linux and macOS systems)ksh(Korn shell)zsh(Z shell)
Think of the fancy point-and-click user-interfaces as running commands on a prompt behind-the-scenes whenever you click something¶
I highly recommend either Linux (Ubuntu, Mint, RedHat) or macOS. The Windows MS-DOS prompt is something else entirely.¶
If you're on a Windows machine, you can either:
- Activate the Ubuntu shell (Windows 10 only) https://msdn.microsoft.com/en-us/commandline/wsl/install_guide
- Install Cygwin https://www.cygwin.com/
- Install VirtualBox ( https://www.virtualbox.org/wiki/VirtualBox ) and run an Ubuntu virtual machine inside
- Go to the computer labs (RedHat or macOS will work)
As of Windows 10, there is now a "bash subsystem" you can enable which is a fully-functional bash command prompt!
Follow these instructions: https://docs.microsoft.com/en-us/windows/wsl/install-win10
Once the subsystem is installed, you can configure it following these instructions: https://docs.microsoft.com/en-us/windows/wsl/initialize-distro
I'd highly recommend this! You'll be able to tinker with the command line through JupyterHub, but it's really nice to have on your own machine.
I have a macOS laptop, an Ubuntu workstation, a bunch of RedHat servers, and a Windows 10 home desktop.
I'm most at home with either macOS or Ubuntu.
It's like learning another language: you'll only get better at it if you immerse yourself in it, even when you don't want to.
Diving in!¶
You've fired up the command prompt (or Terminal in macOS). How do you see what's in the current folder?
Last login: Mon Jan 9 18:36:07 on ttys006 example1:~ squinn$ ls Applications Dropbox Music SpiderOak Hive Desktop Google Drive Pictures metastore_db Documents Library Programming nltk_data Downloads Movies Public rodeo.log example1:~ squinn$
ls¶
Allows you to view the contents of the current directory--folders and files.
But how do we tell the difference between the two? Use an optional -l flag.
(aside: "flags" are options to commands that slightly tweak their behavior to account for different user intentions--like "quit" versus "force quit")¶
example1:~ squinn$ ls -l total 264 drwx------ 7 squinn staff 238 Oct 23 2015 Applications drwx------+ 59 squinn staff 2006 Jan 9 17:49 Desktop drwx------+ 20 squinn staff 680 Dec 23 09:35 Documents drwx------+ 5 squinn staff 170 Jan 9 18:27 Downloads drwx------@ 17 squinn staff 578 Jan 8 18:03 Dropbox drwx------@ 49 squinn staff 1666 Jan 4 15:47 Google Drive drwx------+ 74 squinn staff 2516 Nov 17 15:06 Library drwx------+ 6 squinn staff 204 May 20 2015 Movies drwx------+ 5 squinn staff 170 Oct 22 2014 Music drwx------+ 18 squinn staff 612 Jul 29 11:31 Pictures drwxr-xr-x 37 squinn staff 1258 Jan 4 15:57 Programming drwxr-xr-x+ 5 squinn staff 170 Oct 21 2014 Public drwx------@ 8 squinn staff 272 Jun 30 2015 SpiderOak Hive drwxr-xr-x 9 squinn staff 306 Sep 17 2015 metastore_db drwxr-xr-x 4 squinn staff 136 Apr 27 2016 nltk_data -rw-r--r-- 1 squinn staff 131269 Jan 9 18:32 rodeo.log example1:~ squinn$
Anything that starts with a d on the left is a folder (or directory), otherwise it's a file.
Ok, that's cool. I can tell what is what where I currently am. ...but wait, how do I even know where I am?
example1:~ squinn$ pwd /home/squinn example1:~ squinn$
pwd¶
Pretty straightforward--stands for Print Working Directory. Gives you the full path to where you are currently working. Not really any other needed optional flags.
Great! Now I know where I am, and what is what where I am. How do I move somewhere else?
This would be the same thing as double-clicking a folder to move into it.
example1:~ squinn$ cd Music/ example1:Music squinn$ ls iTunes example1:Music squinn$
You'll notice the output of the ls command has now changed, which hopefully isn't surprising.
Since we've Changed Directories with the cd command--you essentially double-clicked the "Music" folder--now we're in a different folder with different contents; in this case, a lone "iTunes" folder.
Folders within folders represent a recursive hierarchy. We won't delve too much into this concept, except to say that, unless you're in the root directory (/ on Linux, C:\ on Windows), there is always a parent directory--the enclosing folder around the folder you are currently in.
This gives rise to a hierarchical structure of folders.
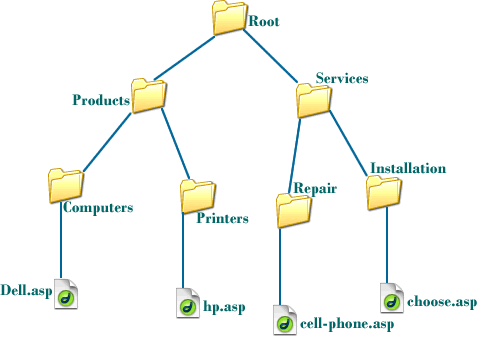
Therefore, while you can always change to a very specific directory by supplying the full path--
example1:~ squinn$ cd /home/squinn/Dropbox example1:Dropbox squinn$ ls Cilia_Papers Imaging_Papers OdorAnalysis Public Computer Case LandUseChange OrNet cilia movies Icon? NSF_BigData_2015 OrNet Videos example1:Dropbox squinn$
--I can also navigate to the parent folder of my current location, irrespective of my specific location, using the special .. notation.
cd ..¶
Takes you up one level to the parent directory of where you currently are.
example1:Dropbox squinn$ pwd /home/squinn/Dropbox example1:Dropbox squinn$ cd .. example1:~ squinn$ pwd /home/squinn example1:~ squinn$
Relative paths vs Absolute paths¶
It's important to distinguish relative versus absolute paths when you're changing directories.
- A relative path is one that is relative (hence, the name) to your current location. The "
.." path is a good example: it means "go to the parent directory of wherever I am now". That "wherever I am now" is the relative part, so typing "cd .." will put you in a different parent directory depending on where you start.
- An absolute path is one that is always the same, no matter where you start. For instance, typing "
cd /home/squinn" will always always take me to the "/home/squinn" folder, no matter where I was when I typed the command.
Relative or Absolute?¶
..
/tmp/file.txt
some/folder/file.txt
Found the pattern yet?
Anything with a preceding / is an absolute path; otherwise, it is considered relative.
Let's see some other examples!
example1: squinn$ ls Lecture1.ipynb example1: squinn$ ls -l total 40 -rw-r--r-- 1 squinn staff 18620 Jan 5 19:54 Lecture1.ipynb example1: squinn$ pwd /home/squinn/teaching/4835/lectures example1: squinn$ cd .. example1: squinn$ pwd
What prints out?
~//home/squinn/home/squinn/teaching/home/squinn/teaching/4835- An Error
$ ls -l total 8 -rw-rw-r-- 1 squinn staff 19 Sep 3 09:08 hello.txt drwxrwxr-x 2 squinn staff 4096 Sep 3 09:08 lecture $ ls *.txt
What prints out?
hello.txt*.txthello.txt lecture- An Error
Spacing Out¶
du - disk usage of files/directores
[squinn tmp]$ du -s 146564 . [squinn tmp]$ du -sh 144M . [squinn tmp]$ du -sh intro 4.0K intro
df - usage of full disk
[squinn tmp]$ df -h . Filesystem Size Used Avail Use% Mounted on pulsar:/home 37T 28T 9.3T 75% /net/pulsar/home
Dude, where's my stuff?¶
From time to time, you'll probably be wondering where you can find certain things.
- locate: find a file system wide. This requires a pre-built database of your filesystem, so you may need to run
updatedbfirst, and every time you install something new. But it's super efficient.
- find: search directory tree. This does an actual manual search of your hard disk every time it runs, so it can be really slow if you search absolutely everything. But consequently it doesn't require rebuilding a database every time you add a new file.
- which: print location of a command. Let's say you're running a command (like
python) and want to know where it's installed on your computer. You can sawwhich pythonand it will tell you.
- man: print manual page of a command. "Manual pages", or "man pages", are the built-in linux manuals for all the commands you could ever need. Just type
man <command>to learn more than you ever cared to know about how to use it!
Save the Environment¶
"Environment variables" are ways of storing little bits of information for you to re-use in your commands. We'll see a version of this in Python, too!
NAME=value: set NAME equal to value No spaces around equals
export NAME=value: set NAME equal to value and make it stick
\$: dereference variable. This is a fancy way of saying "use this variable in something useful." As an example:
$> X=3 $> echo $X 3 $> X=hello $> echo $X hello $> echo X X
Getting at your variables¶
Which does not print the value of X?
echo $Xecho ${X}echo '$X'echo "$X"
It's a little tricky! The key is to notice the little "backtick" operators--these things "`"--that enclose the command you want to run.
Then mentally substitute that command in for its variable, and hopefully you kinda see how it works.
Your Environment¶
env list all set environment variables (the variables we defined from before)
PATH where shell searches for commands. It is VERY IMPORTANT that you leave this variable alone unless you know what you're doing. Although it's kind of inevitable that everyone who uses the command line nukes this variable accidentally at least once...
LD_LIBRARY_PATH library search path (don't worry about this too much)
PYTHONPATH where python searches for modules (also don't worry about this, especially if you just use JupyterHub for everything)
.bashrc initialization file for bash - set PATH etc. This is like a "startup" file that's run every time you open the command prompt, so if there are any variables you want to take on specific values every single time you open up a terminal, this is the place to set them.
History¶
After you've been using the terminal for awhile, you may want to look at all the commands you've run! Or perhaps it took you awhile to figure out exactly how to run a command, and a few days/weeks/months later you find you need to run that command again. There are ways of looking into your command history:
history show commands previously issued
up arrow cycle through previous commands
Ctrl-R search through history for command AWESOME
.bash_history file that stores the history
HISTCONTROL environment variable that sets history options: ignoredups
HISTSIZE size of history buffer
Shortcuts¶
Some clever shortcuts on the command line. By definition, they're not necessary, but they can make your work more efficient.
Tab autocomplete
Ctrl-D EOF/logout/exit
Ctrl-A go to beginning of line
Ctrl-E go to end of line
alias new=cmd. This is a fun one: if you don't like how certain commands are named, you can make up your own!
make a nickname for a command $ alias l='ls -l' $ alias $ l total 40 -rw-r--r-- 1 squinn staff 18620 Jan 5 19:54 Lecture1.ipynb
which is the output of ls -l that we saw earlier!
Commands¶
The first word you type is the program you want to run. bash will search PATH for an appropriately named executable and run it with the specified arguments.
Some example commands we'll be using a lot:
- ipython - start interactive python shell (more later)
- ssh hostname - connect to hostname
- passwd - change your password
- nano - a user-friendly text editor
ssh into jupyterhub.cs.uga.edu and change your password¶
Part 2: Text Manipulation¶
First, a quick review: some of the commands we just covered that will be coming back here.
- ls - list files
- cd - change directory
- pwd - print working (current) directory
- .. - special file that refers to parent directory
- . - the current directory
- cat file - NEW COMMAND: prints out the contents of the file
- more file - NEW COMMAND: gently prints out the contents of a file
I/O Redirection¶
"I/O" is a bit of technical slang for "input/output". It refers to the things that go into a program, and the things that come back out.
For example, the command echo "Hello, world!" can be considered a program--"echo"--with input "Hello, world!". It's just a type of program where the input and output are the same.
With the command line, we therefore refer to specific concepts known as standard input and standard output. Don't worry, this is pretty straightforward:
- Standard input means anything you've typed on the keyboard, and
- Standard output is anything that's printed back inside the main command prompt window
So when you type echo "Hello, world!", the input is provided through standard input (since you typed it), and the output is sent through standard output (since it appeared in the command prompt right below the command you typed).
We can, if we want, redirect those inputs and outputs. For example: if we have a long sequence of programs, perhaps we want the standard output of one program to be the input to the next one.
> send standard output to file
$ echo Hello > h.txt
>> append to file
$ echo World >> h.txt
A quick exercise!
$ echo Hello > h.txt $ echo World >> h.txt $ cat h.txt
What prints out?
- Hello
- World
- HelloWorld
Hello
World- An Error
A slightly different example:
$ echo Hello > h.txt $ echo World > h.txt $ cat h.txt
What prints out?
- Hello
- World
- HelloWorld
Hello
World- An Error
Pipes¶
A pipe (|) redirects the standard output of one program to the standard input of another. It's like you typed the output of the first program into the second. This allows us to chain several simple programs together to do something more complicated.
wc is a very useful little command for "word counting".
$ wc h.txt 1 2 14 h.txt
This means the text in h.txt has 1 line, 2 words, and 14 letters total.
But rather than feed the text into a file first, we could echo it and pipe it directly into the wc command as input:
$ echo hello, world! | wc
1 2 14
Same output! But no filename, because we skipped that step.
Simple Text Manipulation¶
A frequent part of computational biology is, sadly, futzing with data in text files. But with a little bit of knowledge of text-processing command line programs, you can do a lot of this without even using something like Python.
Some programs we've seen:
cat: dump file to stdoutmore: paginated output (a nicer version ofcat, basically)head: show first 10 linestail: show last 10 lineswc: count lines/words/characters
Some new ones:
sort: sort file by line and print out (-n for numerical sort)uniq: remove adjacent duplicates (-c to count occurances)cut: extract fixed width columns from file
Here's a fun example that ties some of these commands up using pipes:
$ cat text a b a b b $ cat text | uniq | wc
What is the first number to print out?
- 1
- 2
- 3
- 4
- 5
- None of the above
A slight wrinkle:
$ cat text a b a b b $ cat text | sort | uniq | wc
What is the first number to print out?
- 1
- 2
- 3
- 4
- 5
- None of the above
Advanced Text Manipulation¶
It's great to be able to dump out text using cat, count words and letters with wc, and sort and uniq-ify the outputs.
But what if I wanted to search a text file for a specific word? Or extract the third word of every line in a file? Or pull out every line that started with the word "The"?
These are all a bit beyond the basic utilities we've described, so we have to explore some more advanced programs.
In particular, we're looking at these three commands:
grep: search contents of file for expressionsed: stream editor - perform substitutionsawk: pattern scanning and processing, great for dealing with data in columns
We'll spend a slide on each one.
grep¶
This searches a file's contents for whatever pattern you specify. The command is organized this way:
pattern can be any collection of letters that you want to match. The lines where the matches are found (if any) are printed to standard out if grep finds any.
grep also has a few flags to change how it works:
-r: recursive search-I: skip over binary files-s: suppress error messages-n: show line numbers-A: show N lines after match-B: show N lines before match
Yay exercise!
$ cat text a b a b b $ grep a text | wc
What is the first number to print out?
- 1
- 2
- 3
- 4
- 5
- None of the above
sed¶
sed is a great search and replace command. If you've ever used "Find and Replace" in Microsoft Word or something like that, this is the command-line version.
It works with this pattern:
sed 's/pattern/replacement/' file
-i: replace in-place (overwrites input file; the default behavior without this flag is to print the changes to standard output and leave the original file as-is)
Basically, I'm searching the file for letters or words that exactly match pattern, and then replacing those with whatever I put in replacement.
So! How about an exercise?
$ sed 's/a/b/' text | uniq | wc
What is the first number to print out?
- 1
- 2
- 3
- 4
- 5
- None of the above
awk¶
This is the single most advanced utility we'll cover for the command line. I'm not overstating to say knowledge and especially mastery of awk gets you jobs.
awk is pretty much a fully-functioning language unto itself, built for scanning, processing, and transforming text data. It's largely used to extract rows or columns of data according to potentially-complex conditions, but it can do just about anything you'd like, really.
It processes a file line-by-line, and if a condition holds true, it runs a simple program on the text in that line.
awk 'optional condition {awk program}' file
- -Fx make x the field deliminator (default whitespace)
- NF number of fields on current line
- NR current record number
- \$0 full line
- \$N Nth field
Before we get to exercises, let's try a few things out.
$ cat names id last,first 1 Smith,Alice 2 Jones,Bob 3 Smith,Charlie
Try these:
$ awk '{print $1}' names
$ awk -F, '{print $2}' names
$ awk 'NR > 1 {print $2}' names
$ awk '$1 > 1 {print $0}' names
$ awk 'NR > 1 {print $2}' names | awk -F, '{print $1}' | sort | uniq -c
Hint: $1, $2, and similar refer to columns in the text (0-indexed!)
Exercises¶
Let's walk through a couple of examples with some real files. You can download these off the course website. In fact, you can even do it from the command line with wget!
mkdir intro cd intro wget https://eds-uga.github.io/cbio4835-fa18/files/Spellman.csv wget https://eds-uga.github.io/cbio4835-fa18/files/1shs.pdb
- How many data points are in Spellman.csv?
- The first three letters of the systematic open reading frames are: 'Y' for yeast, the chromosome number, then the chromosome arm. In the dataset, how many ORFs from chromosome A are there?
- How many are there from each chromosome?
- each chromosome arm?
- How many data points start with a positive expression value?
- What are the 10 data points with the highest initial expression values?
- Lowest?
- How many lines are there where expression values are continuously increasing for the first 3 time steps?
- Sorted by biggest increase?
wc Spellman.csv (gives number of lines, because of header this is off by one)
grep YA Spellman.csv |wc
grep ^YA Spellman.csv |wc (this is a bit better, ^ matches begining of line)
grep ^YA -c Spellman.csv (grep can provide the count itself)
awk -F, 'NR > 1 {print $1}' Spellman.csv | cut -b 1-2 | sort | uniq -c
awk -F, 'NR > 1 {print $1}' Spellman.csv | cut -b 1-3 | sort | uniq -c
awk -F, 'NR > 1 && $2 > 0 {print $0}' Spellman.csv | wc
awk -F, 'NR > 1 {print $1,$2}' Spellman.csv | sort -k2,2 -n | tail
awk -F, 'NR > 1 {print $1,$2}' Spellman.csv | sort -k2,2 -n -r | tail
awk -F, 'NR > 1 && $3 > $2 && $4 > $3 {print $0}' Spellman.csv |wc
awk -F, 'NR > 1 && $3 > $2 && $4 > $3 {print $4-$2,$0}' Spellman.csv | sort -n -k1,1
More Exercises¶
- Create a pdb file from 1shs that consists of only ATOM records.
- Create a pdb with only ATOM records from chain A.
- How many carbon atoms are in this file?
grep ^ATOM 1shs.pdb > newpdb.pdb (^matches beginning of line)
grep ^ATOM 1shs.pdb | awk '$5 == "A" {print $0}'
#this is UNSAFE with pdb files since there is no guarantee that fields
#will be whitespace seperated, safer is:
grep ^ATOM 1shs.pdb | awk ' substr($0,22,1) == "A" {print $0}' > newpdb.pdb
grep ^ATOM 1shs.pdb | awk ' substr($0,22,1) == "A" {print $0}' | cut -b 78- | sort | uniq -c
Administrivia¶
- Did everyone finish the pre-test? It was due today before lecture. https://docs.google.com/forms/d/1ka9yH5G3bOCfdJUTaeZXV2BdtvqqsiPaxnvKI2f4YK4/
- JupyterHub is up and running! Check to make sure that you can log in (instructions were posted in Slack).
- Assignment 1 will also be released later today. Due in two weeks!
Resources¶
- A BASH cheatsheet: http://eds-uga.github.io/cbio-x835-sp20/notes/bash_cheatsheet.pdf