Lecture 18: Introduction to Computer Vision¶
CBIO (CSCI) 4835/6835: Introduction to Computational Biology
Overview and Objectives¶
This week, we're moving into image processing. In this lecture, we'll touch on some core concepts around computer vision, or the field dedicated to machine understanding of images. By the end of this lecture, you should be able to
- Read in and display any image using Python
- Understand the basic components and core data structures of images
- Describe core image processing techniques such as thresholding, equalization, and autocontrast
- Recall some of the computer vision packages available in Python for more advanced image processing
Part 1: Computer Vision¶
Whenever you hear about or refer to an image analysis task, you've stepped firmly into territory occupied by computer vision, or the field of research associated with understanding images and designing algorithms to do the same.

Examples of Computer Vision¶
You can probably name numerous examples of computer vision already, but just to highlight a couple:

- Facebook and Google use sophisticated computer vision methods to perform facial recognition scans of photos that are uploaded to their servers. You've likely seen examples of this when Facebook automatically puts boxes around the faces of people in a picture, and asks if you'd like to tag certain individuals.
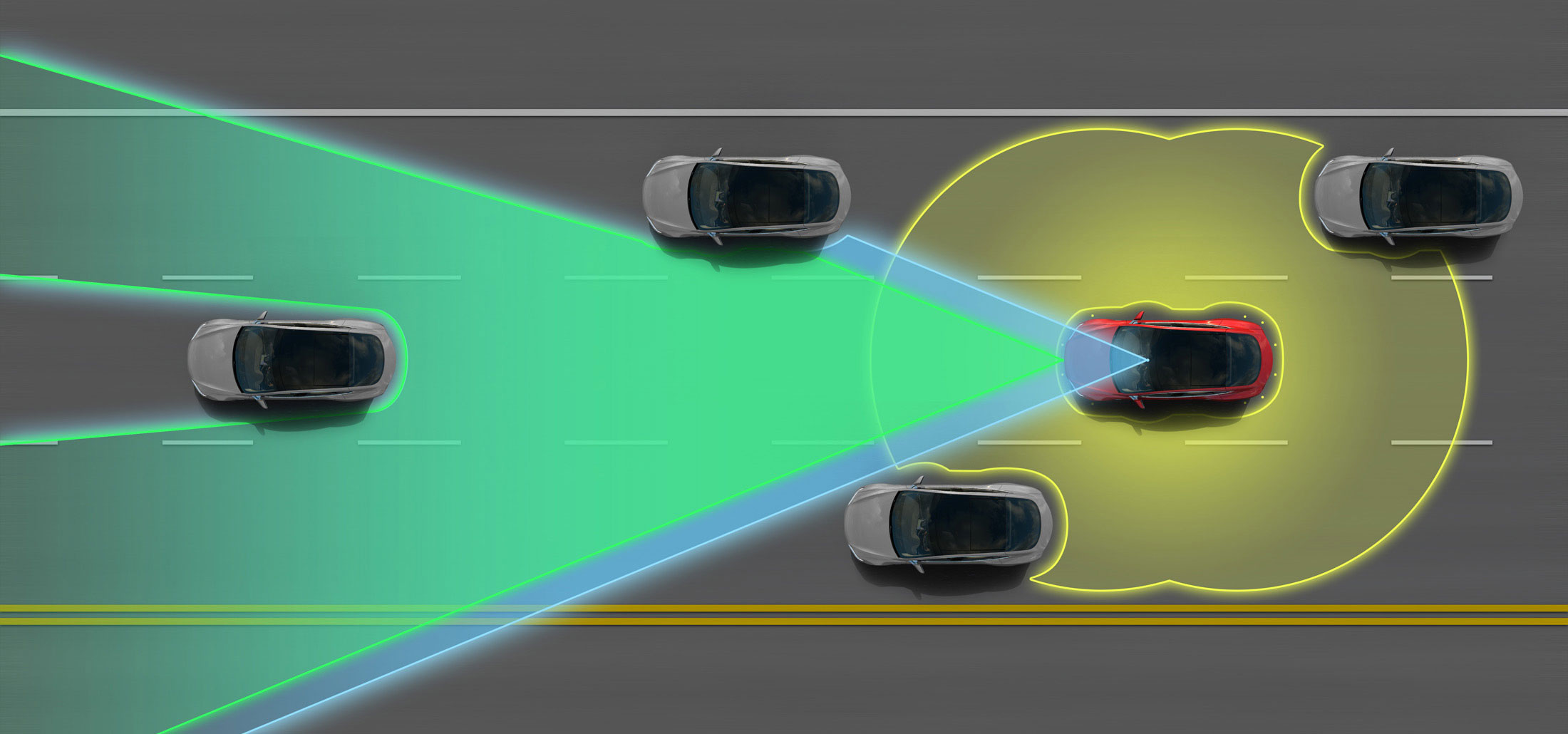
- Tesla Motors' "Autopilot" and other semi-autonomous vehicles use arrays of cameras to capture outside information, then process these photos using computer vision methods in order to pilot the vehicle. Google's experimental self-driving cars use similar techniques, but are fully autonomous.
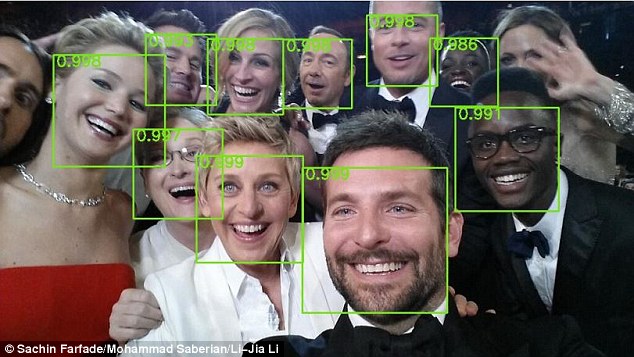
- The subarea of machine learning known as "deep learning" has exploded in the last five years, resulting in state-of-the-art image recognition capabilities. Google's DeepMind can recognize arbitrary images to an extraordinary degree, and similar deep learning methods have been used to automatically generate captions for these images.
This is all to underscore: computer vision is an extremely active area of research and application!
- Automated categorization and annotation of YouTube videos (identification of illegal content?)
- Analyzing photos on your smartphones
- License plate and facial recognition for law enforcement officials
- Disabled access to web technologies
- Virtual reality
Images and their Representations¶
From the perspective of the computer, the simplest constituent of an image is a pixel.
- pix: picture
- el: element
A pixel is a picture element.
- In a grayscale image, the pixel contains the intensity. Depending on the image format this may range from 0-1, 0-255, or be any floating point number.
- In a color image, a pixel is (usually) a triple (red, green, blue) of color values where each color intensity ranges from 0-255 (24-bit color).
(There are many other image formats and representations, but they tend to be variations on this theme)
In either grayscale or color, the pixels are arranged in rectangular arrays, one for each color channel (1 for grayscale, 3 for RGB).
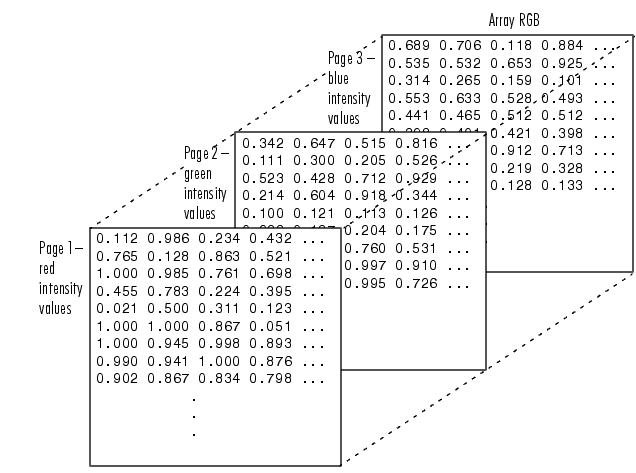
(What could these arrays possibly be in Python?)
Part 2: Loading and Manipulating Images¶
Let's jump in and get our hands dirty! First, let's use a relevant image:
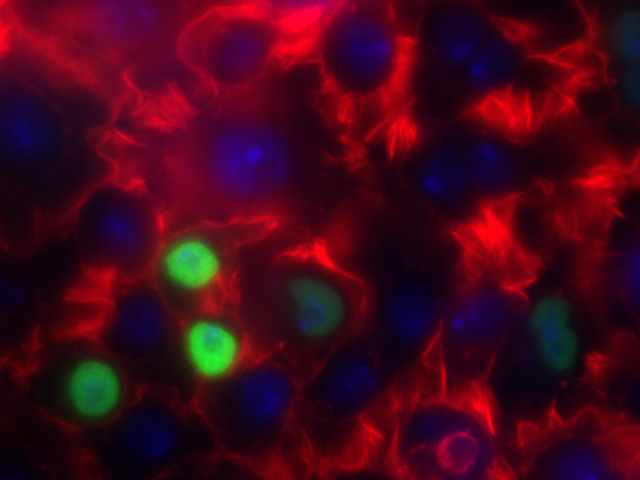
- Actin
- HSP27
- DAPI
I've stored this image in the course GitHub repository under lectures/ComputerVision ( https://github.com/eds-uga/cbio4835-fa18 ) if you're interested.
Here's how to load the images in Python:
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
# Loads the image (just like a text file!)
img = mpimg.imread("ComputerVision/image1.png")
print(type(img))
<class 'numpy.ndarray'>
Just a regular NumPy array!
Let's see if we can visualize it.
plt.imshow(img)
<matplotlib.image.AxesImage at 0x11b27b9b0>
This shows the whole image, all three channels.
print(img.shape)
(480, 640, 3)
As evidenced by the .shape property of the NumPy array, there are three dimensions to this image:
- the first is height (or rows)
- the second is width (or columns)
- the third is color (or depth)
Each slice of the third dimension is a color channel, of which there are 3: one for red, one for green, and one for blue (hence: RGB).
We can plot them separately!
# First, separate out the channels.
r = img[:, :, 0]
g = img[:, :, 1]
b = img[:, :, 2]
# Now, plot each channel separately.
f = plt.figure(figsize = (12, 6))
f.add_subplot(1, 3, 1)
plt.imshow(np.array(r), cmap = "gray")
f.add_subplot(1, 3, 2)
plt.imshow(np.array(g), cmap = "gray")
f.add_subplot(1, 3, 3)
plt.imshow(np.array(b), cmap = "gray")
<matplotlib.image.AxesImage at 0x11c011710>
Image analysis of any kind is usually done on a single channel.
Since images are stored as NumPy arrays, all the usual NumPy functionality (besides slicing, as we saw earlier) is available to you.
- Want to compute the maximum and minimum pixel values in the images?
print(np.max(img))
print(np.min(img))
1.0 0.0
- Want to compute the average and median pixel values?
print(np.mean(img))
print(np.median(img))
0.13152441 0.07058824
- How about the median of each of the red, green, and blue channels separately?
print(np.median(r))
print(np.median(g))
print(np.median(b))
0.20784314 0.007843138 0.07450981
Converting Image Types¶
Recall that our img object was loaded from a PNG image; this is the only format type that Matplotlib natively supports (more on that later).
When you read an image into Python, it will automatically detect the format and read it into the closest approximate Python data format it can. However, you can always manually convert it once it's in Python.
For instance, we use a slightly different approach to instead read in our image as grayscale:
import scipy.ndimage as ndimg
img_gray = ndimg.imread("ComputerVision/image1.png", flatten = True) # The "flatten" arg is critical
print(img_gray.shape)
(480, 640)
/opt/python/lib/python3.6/site-packages/ipykernel_launcher.py:3: DeprecationWarning: `imread` is deprecated! `imread` is deprecated in SciPy 1.0.0. Use ``matplotlib.pyplot.imread`` instead. This is separate from the ipykernel package so we can avoid doing imports until
Note how there are only 2 dimensions now--just a height and width.
There is no need for a 3rd dimension because there's only 1 channel: luminescence, or grayscale intensity.
plt.imshow(img_gray, cmap = "gray")
<matplotlib.image.AxesImage at 0xb1df891d0>
We can access individual pixels, just as you would individual elements of a matrix NumPy array (because that's all it is):
print(img_gray[100, 200])
35.139
print(img_gray[150, :])
[11.606 12.503 11.279 11.692 11.692 11.621 12.208 13.034 12.518 12.023 12.148 11.436 12.148 12.023 11.963 12.479 13.305 12.892 12.821 12.935 14.43 12.967 13.576 13.206 13.178 13.662 14.26 14.63 13.733 15.402 13.619 13.75 13.576 14.718 14.305 12.593 11.468 12.909 13.806 12.909 12.98 13.578 12.752 12.823 11.513 13.41 11.926 12.812 13.068 12.78 11.769 11.829 13.21 12.808 12.982 11.911 12.384 13.053 13.053 13.961 13.064 14.858 15.146 17.549 18.435 16.527 17.054 16.228 18.136 17.848 17.239 17.609 18.136 18.136 15.929 18.435 15.929 17.239 16.94 16.342 17.125 16.527 15.755 19.631 17.538 18.25 18.848 18.549 20.044 21.539 22.436 21.849 24.643 27.334 29.911 33.613 37.201 38.984 34.923 37.614 36.119 36.407 35.336 36.005 36.532 34.749 35.222 35.934 33.841 34.151 36.244 36.646 36.956 38.554 39.647 37.853 40.06 41.854 41.854 43.833 43.278 40.245 41.854 39.288 39.576 41.185 36.7 37.184 39.092 36.928 37.412 35.917 36.216 33.949 34.846 34.547 33.949 34.351 32.878 34.362 34.96 34.96 37.238 35.672 33.65 36.156 34.672 35.373 34.672 33.106 34.074 37.292 36.58 35.797 37.096 38.374 35.313 35.313 36.993 35.199 36.438 39.015 40.51 42.005 41.179 39.385 40.624 41.407 38.912 38.118 40.51 36.808 37.705 37.221 36.922 39.129 39.727 40.396 41.706 42.603 40.211 40.026 41.407 39.026 40.635 38.542 40.934 39.325 38.357 38.243 38.656 41.934 38.531 37.748 35.666 37.036 35.965 34.47 34.698 34.171 33.502 33.573 34.399 33.317 31.452 30.669 30.854 32.577 32.62 30.282 32.293 30.738 29.759 30.041 31.269 29.529 30.458 29.686 30.914 30.783 29.804 30.157 31.385 30.428 28.634 29.346 29.944 28.139 30.944 30.628 29.133 30.486 29.208 29.789 29.604 30.615 28.451 32.723 30.886 30.516 30.445 30.815 29.234 30.419 30.359 29.576 29.391 31.157 30.047 30.987 33.138 34.007 36.001 32.484 33.452 33.837 35.788 34.663 32.598 32.626 30.404 29.89 27.554 26.044 26.855 27.709 27.182 27.296 26.057 27.552 24.704 24.177 24.917 24.988 23.407 23.962 22.467 22.196 22.381 21.783 21.327 20.985 20.871 21.056 20.714 19.589 18.051 19.275 19.047 17.58 16.537 16.113 17.494 15.13 15.429 14.614 16.408 16.098 14.984 15.158 15.158 14.158 15.528 15.653 13.25 16.126 15.941 14.517 16.724 15.713 16.507 16.083 17.393 14.588 15.61 16.393 16.083 14.3 18.29 15.784 17.094 19.187 17.888 16.681 19.084 19.372 18.66 17.877 18.073 18.66 17.361 17.578 16.453 17.959 17.66 18.144 17.649 19.927 18.742 19.742 20.052 21.308 22.145 27.103 27.228 27. 30.588 32.311 35.475 35.187 37.568 39.373 39.857 39.971 42.548 43.445 41.352 45.043 45.538 45.641 47.147 47.517 45.951 40.754 38.362 31.485 31.599 25.505 24.722 21.732 17.916 15.638 14.143 13.246 12.947 13.246 11.637 12.05 13.05 13.545 14.442 14.143 16.834 16.834 19.411 21.02 22.216 30.588 37.454 45.353 58.694 67.664 59.879 54.095 42.548 32.371 31.186 31.186 32.855 33.991 32.083 34.774 35.671 35.258 33.393 35.774 35.486 35.062 35.073 31.898 25.32 22.031 22.743 22.743 22.02 20.166 19.269 18.845 17.888 19.443 17.578 16.268 15.012 14.887 13.816 12.919 14.218 13.022 12.734 13.549 10.57 10.086 11.994 10.613 10.684 11.499 9.83 11.439 9.944 9.759 10.172 9.389 10.813 9.987 9.503 9.019 9.432 11.514 9.731 8.823 9.318 9.389 7.666 8.748 10.417 9.602 8.694 9.004 10.086 9.488 9.901 10.303 10.189 10.015 9.107 10.232 9.862 8.791 8.905 10.372 11.198 10.714 10.344 10.686 12.523 12.723 12.869 12.738 13.493 14.319 15.487 15.188 15.302 14.633 15.388 15.958 16.528 17.24 16.642 16.485 16.442 17.253 15.259 15.7 13.706 14.332 14.218 14.289 14.616 13.708 11.953 12.622 12.693 10.813 12.977 12.923 12.293 13.56 12.549 13.147 11.837 12.62 12.549 11.424 12.321 12.506 12.99 12.392 13.093 12.093 13.474 11.794 12.164 12.463 11.74 12.05 10.854 11.153 12.648 11.751 13.616 12.947 13.73 12.833 12.833 14.328 11.936 14.029 13.915 12.534 12.534 14.616 13.018 14.926 12.121 9.843 11.224 8.832 10.925 11.338 12.42 12.719 14.029 13.616 12.833 12.719 12.833 15.111 13.502 13.018 14.029 15.709 13.616 14.513 14.997 15.524 14.812 15.111 16.307 16.606 16.606 17.019 16.008 15.41 16.606 17.204 17.802 20.792 18.4 18.514 20.379 22.575 22.885 22.39 23.369 25.75 24.266 28.256 28.267 26.049 25.266 26.163 24.07 26.348 26.359 25.75 25.75 25.875 26.049 25.761 28.256 27.071 26.473 28.555 28.267 31.246 31.545 30.958 30.463 26.761 26.462 28.555 25.451 26.946 25.462 25.864 23.657 22.874 21.39 20.493 18.873 19.297 19.471 17.976 18.275 18.09 19.286 18.389 19.172 18.574 19.77 21.265 20.667 19.172 22.151 23.76 26.348 29.74 30.822 29.441]
print(np.max(img_gray[:, 400]))
78.428
If you so desire, you can even modify the pixel values directly, again just as you would for a regular NumPy array.
Fair warning: doing this alters the image! You may want to copy the image structure first...
for i in range(img_gray.shape[0]):
for j in range(120, 130):
img_gray[i, j] = 255
plt.imshow(img_gray, cmap = "gray")
<matplotlib.image.AxesImage at 0xb1dff2860>
Histograms¶
Another very useful way of obtaining information about an image is to view the histogram of pixel values.
You can do this regardless of whether it's a grayscale or RGB image, though in the latter case it's useful to plot the pixel values separated by channel.
First, let's re-import the image as grayscale and take a look at how the pixel values show up in a histogram:
img_gray = ndimg.imread("ComputerVision/image1.png", flatten = True)
_ = plt.hist(img_gray.flatten(), bins = 25)
/opt/python/lib/python3.6/site-packages/ipykernel_launcher.py:1: DeprecationWarning: `imread` is deprecated! `imread` is deprecated in SciPy 1.0.0. Use ``matplotlib.pyplot.imread`` instead. """Entry point for launching an IPython kernel.
This tells us some very useful information--primarily, that most of the pixel values are centered around what seems like a pretty low number (20-30), so by and large the image is very dark (which we saw).
There do seem to be a few light spots on an island around 120-140, but that's it.
Let's take a look now at each channel individually.
fig = plt.figure(figsize = (16, 4))
plt.subplot(131)
_ = plt.hist(r.flatten(), bins = 25, range = (0, 1), color = 'r')
plt.subplot(132)
_ = plt.hist(g.flatten(), bins = 25, range = (0, 1), color = 'g')
plt.subplot(133)
_ = plt.hist(b.flatten(), bins = 25, range = (0, 1), color = 'b')
Recall what each channel represented:
- Actin
- HSP27
- DAPI
There seems to be very little HSP27, while there is tons of actin and the quantity of DAPI falls somewhere in between.
...oh wait, did you see the scales for each one?
x = np.linspace(0, 1, 25)
plt.plot(x, np.histogram(r.flatten(), bins = 25)[0], color = 'r', label = 'Actin')
plt.plot(x, np.histogram(g.flatten(), bins = 25)[0], color = 'g', label = 'HSP27')
plt.plot(x, np.histogram(b.flatten(), bins = 25)[0], color = 'b', label = 'DAPI')
plt.legend()
<matplotlib.legend.Legend at 0xb1e76f320>
So, yes:
- Clearly, very little HSP27 signal, relative to the other stains. Most of those pixels are black (0).
- There does seem to be a decent amount of DAPI signal, but like HSP27 it too drops off very quickly; not many DAPI pixels with brightness greater than 0.4 or so.
- The actin signal is probably the most interesting one, in that it's very diffuse--very few black (0) pixels, no white pixels either (1), but somewhere in between for the most part. This is fairly characteristic for actin.
Equalization¶
While we're on the topic of histograms, there is a convenient way to try and "reshape" the pixel histograms so as to make the resulting image a bit sharper. This is called histogram equalization.
The idea is simple enough: re-map the pixel values in the image so that the corresponding histogram is perfectly flat.
Basically it tries to fill in the "valleys" and flatten the "peaks" of the pixel histograms we saw earlier--this has the effect of bringing out very dim signal and dampening oversaturated signal.
Let's see an example, using one of the image channels.
from PIL import Image, ImageOps
img_pil = Image.open("ComputerVision/image1.png")
beq = ImageOps.equalize(img_pil.split()[2])
f = plt.figure(figsize = (12, 6))
f.add_subplot(1, 2, 1)
plt.imshow(b, cmap = 'gray')
f.add_subplot(1, 2, 2)
plt.imshow(np.array(beq), cmap = 'gray')
<matplotlib.image.AxesImage at 0xb1e6094a8>
We can directly see why these two images look different (and, specifically, what histogram equalization did) by recomputing the channel histograms:
plt.plot(img_pil.split()[2].histogram(), 'b')
plt.plot(beq.histogram(), 'k')
[<matplotlib.lines.Line2D at 0xb1e575978>]
Autocontrast¶
Autocontrast is another tool that modifies the pixel histograms to try and make the resulting images more viewable. In this case, the goal of autocontrast is to maximize (normalize) image contrast.
This function calculates a histogram of the input image, removes cutoff percent of the lightest and darkest pixels from the histogram, and remaps the image so that the darkest remaining pixel becomes black (0), and the lightest becomes white (255).
In essence, you choose some percentage cut-off (say: 50%, or 0.5), removes that fraction of pixels that are both darkest and lightest (assumes they're noise and throws them away), then remaps the remaining pixels.
Here's what it might look like:
bcon = ImageOps.autocontrast(img_pil.split()[2], 0.5)
f = plt.figure(figsize = (12, 6))
f.add_subplot(1, 2, 1)
plt.imshow(b, cmap = "gray")
f.add_subplot(1, 2, 2)
plt.imshow(np.array(bcon), cmap = "gray")
<matplotlib.image.AxesImage at 0xb1e9c19e8>
In this case, we're trying to chop off pixel values at both ends of the histogram (lightest and darkest) and reshuffling the others around to make them more visible, hopefully improving contrast.
The effects on the underlying histograms look like:
plt.plot(img_pil.split()[2].histogram(), 'r')
plt.plot(bcon.histogram(), 'k')
[<matplotlib.lines.Line2D at 0xb1e575cf8>]
It closely mimics the original histogram, but because some values at the tails were thrown away, all the other values were reshuffled--you end up with more pixels some of the middle values, which is (presumably) the signal you're interested in.
Thresholding¶
Thresholding is the process by which you define a pixel threshold--say, the value 100--and set every pixel below that value to 0, and every pixel above that value to 255.
In doing so, you binarize the image, as each pixel takes on only one of two possible values.
Remember boolean indexing?
(head on over to Lecture 6 if you're a little fuzzy on the details)
In short, you can create masks based on certain boolean conditions so you can modify certain parts of the array while holding the others constant.
Here's the example straight from the lecture:
x = np.random.standard_normal(size = (7, 4))
print(x)
[[ 0.52596627 -2.88941486 0.09271166 -0.21067344] [-0.12179478 -1.05645342 -0.12380148 -0.24558527] [-0.27362056 0.03955863 1.46031891 -0.59451959] [-0.54946308 0.11931622 -0.89393382 -0.37672314] [ 0.23949055 1.17503008 1.25029635 -0.54126771] [-1.38006784 -1.45153849 1.57108929 0.21122822] [ 0.32417432 -0.51411142 0.34512795 0.07949588]]
If we just want the positive numbers, we can define a mask using the condition you'd find in an if statement:
mask = x < 0 # For every element of x, ask: is it < 0?
print(mask)
[[False True False True] [ True True True True] [ True False False True] [ True False True True] [False False False True] [ True True False False] [False True False False]]
The mask is just a bunch of True and False values.
Now we can use the mask to modify the parts of the original array that correspond to True in the mask:
x[mask] = 0.0
print(x)
[[0.52596627 0. 0.09271166 0. ] [0. 0. 0. 0. ] [0. 0.03955863 1.46031891 0. ] [0. 0.11931622 0. 0. ] [0.23949055 1.17503008 1.25029635 0. ] [0. 0. 1.57108929 0.21122822] [0.32417432 0. 0.34512795 0.07949588]]
Back to images! Let's use a threshold on our blue channel:
b_thresh = np.array(bcon) > 120 # Every pixel greater than 120 is "True", otherwise it's "False"
f = plt.figure(figsize = (12, 6))
f.add_subplot(1, 2, 1)
plt.imshow(np.array(bcon), cmap = "gray")
f.add_subplot(1, 2, 2)
plt.imshow(b_thresh, cmap = "gray")
<matplotlib.image.AxesImage at 0xb1eb4fac8>
Any ideas how we might, say, count the number of cells?
Part 3: Computer Vision in Python¶
There is an entire ecosystem of computer vision packages for Python.
Some are very general (a lot like scipy.ndimage and PIL) while some are very specific to certain classes of problems.
You could spend an entire career with just one or two of these packages, but very briefly I'll name a few of the most popular.
(We'll make use of some of them!)
scikit-image¶

If scipy.ndimage or PIL proves to be insufficient for your needs, this should be the first stop you take in looking for alternatives.
It has a wealth of general-purpose image processing routines built-in. It's actively developed and very easy to use, and integrates well with NumPy and SciPy.
It also comes with a bunch of basic tutorials and sample data to help you get your feet wet.
mahotas¶
This is another excellent general-purpose image processing library, though it has a slight preference for bio-imaging applications. After all, its author is a computational biologist!
Like scikit-image, it's actively developed, easy to use, and integrates fully with the NumPy + SciPy scientific computing environment for Python.
This is probably your first stop if you're looking for some basic bioimaging tools.
OpenCV¶

OpenCV (for "Open Computer Vision") is the Grand Daddy of image processing packages.
You'll want to use this if computer vision is a significant part of your day-to-day career. It's not for the faint of heart, however: it's a C++ library with Python bindings, which means you have to install from source, and that can be painful depending on how (un)comfortable you are with compiling things from scratch.
(though if you use the Anaconda distribution of Python, and you connect it to the conda-forge channel, you can download pre-built OpenCV packages that WAY SIMPLIFY this process)
That said, OpenCV has everything:
- automated image segmentation
- facial recognition
- video stabilization
- optical flow algorithms
- image stitching
- filtering
- warping
- matching
- deep learning
- ...
The list goes on and on.
It's well-maintained, well-documented, and while it can be a little tricky to use, it has a huge community of developers and users ready to help.
Like scikit-image, it also provides a ton of tutorials for typical use-cases, though OpenCV's definition of "typical" is a little different: they're actually pretty in-depth!
Administrivia¶
- Assignment 4 is due today!
- Project proposals are also due today!
Additional Resources¶
- Matplotlib image tutorial http://matplotlib.org/users/image_tutorial.html
- scikit-image http://scikit-image.org/
- mahotas http://mahotas.readthedocs.io/en/latest/
- OpenCV http://opencv.org/
- OpenCV Python tutorials http://docs.opencv.org/3.2.0/d6/d00/tutorial_py_root.html