Lecture 22: Open Science¶
CBIO (CSCI) 4835/6835: Introduction to Computational Biology
Overview and Objectives¶
"Open Science" is something of an umbrella term encompassing everything related to reproducible, transparent science.
There have been some complaints that Open Science is amorphous and ambiguous, that its prescription for reproducibility is not, in itself, reproducible.
However, Open Science is broadly defined and meant to appeal to every area of science, from life sciences to computational sciences to theoretical sciences. What Open Science looks like is field-specific, but there are general principles that cut across all fields of science.
By the end of this lecture, you should be able to
- Define "open science" and its importance to scientific inquiry
- Recall the core strategies for reproducing and replicating results
- Package and release your Python code via distribution channels
Part 1: What is "Open Science"?¶

Simply put, Open Science is the movement to make all scientific data, methods, and materials accessible to all levels of society.
Why is this a good thing?
- The vast majority of research is publicly funded; it would seem logical that the public have access to it!
- Open and transparent research makes peer review and replication much easier.
- There is some convincing evidence that Open Science gives projects more downstream impact in the scientific community
Nonetheless, there are some downsides to making everything openly available.
- The deluge of science will overwhelm already heavily-burdened researchers.
- The tools could be used for more nefarious purposes (a good example is a particularly virulent strain of influenza that researchers were experimenting with a few years back that could potentially be used as a bioweapon).
My opinion--as you've probably guessed by the title of the lecture--is that the benefits of good Open Science practices outweigh the drawbacks, for the following reason:
I learn best when I can dig in and get my hands dirty.
Reading a paper or even a blog post that vaguely describes a method is one thing. Actually seeing the code, changing it, and re-running it to observe the results is something else entirely and, so I believe, is vastly superior in educational terms.
The scientific deluge is legitimate, though this was already happening even without the addition of open data, open access, and open source. And it would seem that, while we do absolutely need to exercise caution in our research and not pursue the ends by any means necessary, science is the pursuit of knowledge for its own sake and that should also be respected to the highest degree.
To that end, there are six main themes that comprise the Open Science guidelines.
- Open data: all data used in the project should be made available.
- Open source: all code written in the project should be publicly available.
- Open methods: the exact procedure of the project is publicly documented.
- Open review: correspondence between reviewers and authors is public.
- Open access: resulting publications are publicly available.
- Open education: all education materials are publicly available.
1: Open Data¶

If you had to pick the "core" of Open Science, this would probably be it. All of the data used in your study and experiments are published online.
This is definitely a shift from prior precedent; most raw data from scientific experiments remain cloistered.
The situation is further complicated by Terms of Service agreements that prohibit the sharing of data collected.
- For example: if you hooked up a Python client to listen to and capture public Twitter posts, you are forbidden from sharing the Twitter data publicly.
- Which seems odd, given that the data are public anyway, but there you go.
Repositories and online data banks have sprung up around this idea. Many research institutions host their own open data repositories, as do some large tech companies.
- CERN, the organization behind the Large Hadron Collider, has posted its data online: http://opendata.cern.ch/about/CMS
- Amazon has released its own set of large public datasets: https://aws.amazon.com/public-data-sets/
- Kaggle also has some pretty fantastic open datasets from its competitions: https://www.kaggle.com/datasets
- DataHub is a general-purpose repository for anyone to submit their own datasets. https://datahub.io/en/dataset
2: Open Source¶
This is probably the part you're most familiar with. Any (and all) code that's used in your project is published somewhere publicly for download.
There are certainly conditions where code can't be fully open sourced--proprietary corporate secrets, pending patents, etc--but to fully adhere to Open Science, the code has to be made completely available for anyone.
Like with open data, there are numerous repositories across the web that specialize in providing publicly-available versioning systems for both maintaining and publishing your code.
- GitHub is easily the most popular, and is where the materials for this lecture are published! https://github.com/

- BitBucket is another option that also uses git to manage team codebases https://bitbucket.org/

- SourceForge is one of the oldest and most well-known online repositories https://sourceforge.net/

3: Open Methods¶
This is probably the trickiest item. How does one make methods reproducible?
Open source code is part of it, but even more important is the effort put into making the methods in the code understandable. This takes several forms:
- Documentation, both as accompanying doc files (e.g. JavaDoc) as well as in-code comments
- Proofs of the methods devised if they're novel, or references to their original sources
- Pre-packaged examples that will run with little or no prior configuration on the user's part
- Pre-registered methods before any work is conducted
- Self-contained virtual container scripts that satisfy all the prerequisites
4: Open Review¶
The cornerstone of the scientific process is that of peer review: your peers, your colleagues, your fellow researchers should vet your work before it's officially included as part of the scientific literature.
However, this process is fraught with ambiguity and opacity. Conflicts of interest can potentially lead to biased reviews (if you're reviewing the paper of a competitor, it isn't exactly in your best interest to go easy on them), and it can be difficult to assess a published paper in the public sphere without a trail of edits from which to begin.
Online open review journals such as The Journal of Open Source Software (JOSS) have begun proliferating to address this shortcoming.
Reviews essentially take the form of GitHub tickets, and researchers in the field can discuss and debate the merits of the work in an open forum.
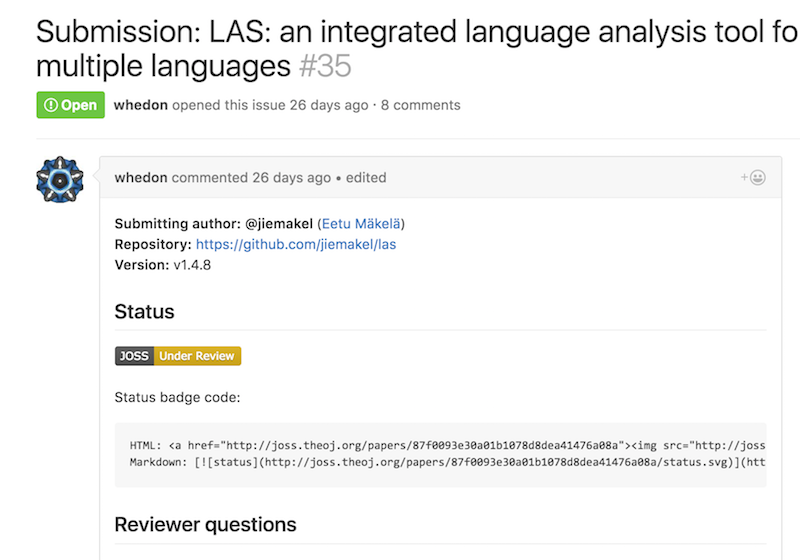
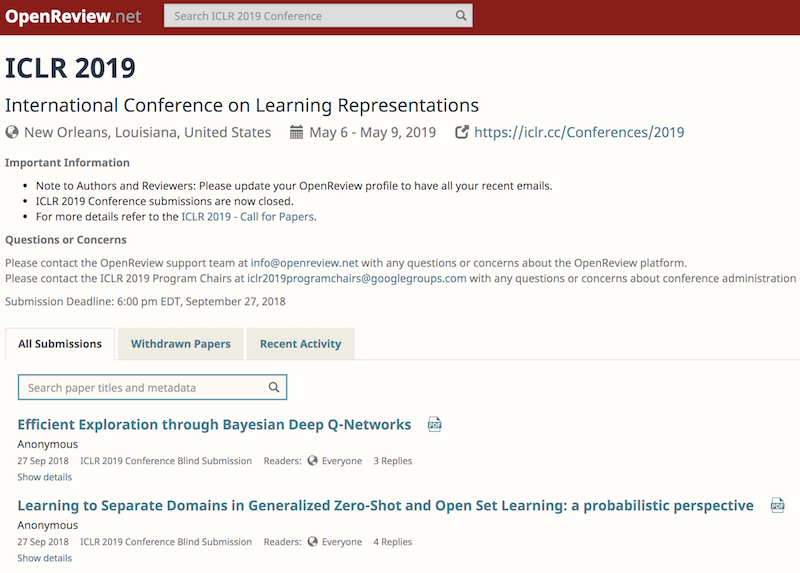
There's also a site, called "Open Review", where conferences can elect to have their papers openly and publicly reviewed, threaded-comment style.
5: Open Access¶
Once a project is published, the paper should be made publicly available for anyone, anywhere to download and read for themselves.
Easily the most popular open access paper repository is arXiv (pronounced "archive"...geddit?). Other repositories modeled directly after its success, such as bioRxiv, have already started springing up.
arXiv is already hugely popular.

In fact, so many papers are archived here on a regular basis, that someone created their own open source "aggregator" service: http://www.arxiv-sanity.com , which collates the papers you want to read and helps filter out all the others.
Preprints have become so popular, that versions of arXiv have popped up for almost every area!

6: Open Education¶
This and Open Methods are closely related. In this sense, any course materials that come from research that is done are made available for others.
MIT OpenCourseWare is probably the best example of open education at work, but these types of sites are proliferating.
- Many courses (including this one!) are making their materials freely available on GitHub.
- Tech companies such as GitHub (aptly named: GitHub Classroom) are developing educational tools specifically aimed at student classrooms. Amazon Web Services provides a similar platform for course materials, often bundled with its cloud compute services.
Note: This does not by default include MOOCs. If MOOCs make their materials freely available online, then and only then would it fall under this section.
So that's all great! But...
What can I do? Where do I start?
Part 2: Open Science Best Practices¶
In the previous section we went over the absolute essentials for an Open Science project or research endeavor. It included some examples of real-world services and tools to help expedite the process.
Here, we'll go into a bit more detail as to exactly how you should tweak your projects to be truly Open Science. It's not really something you can just "tack on" at the end; rather, you'll need to design your projects from the onset to be part of the Open Science initiative.
When you invent the next TensorFlow, we'll all be thanking you for following these best practices!
Version control¶
This is a good starting point for any project.
Version control keeps track of changes within your code base.
It's a lot like Google docs, but for code.
You all know Dropbox? Has it ever saved your life? If so, the next time you start a coding project, use some form of version control.
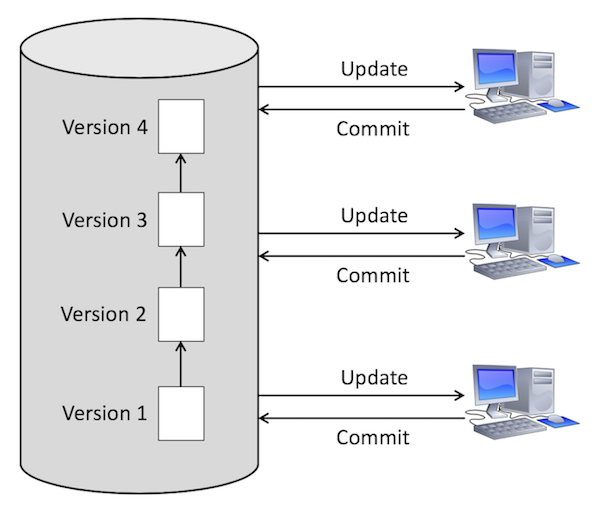
There are several different version control systems out there for code, depending on what you like.
I personally prefer git, but I've used all of these before.

Anatomy of an open source project¶
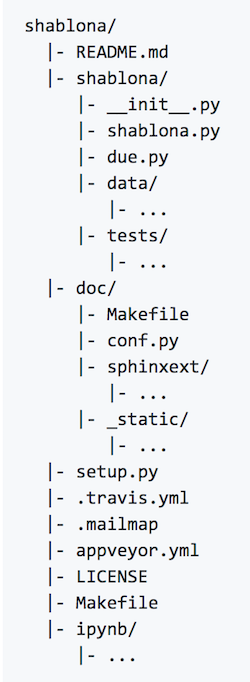
There are some common files and folder hierarchies to implement at the very start of a new project. The shablona GitHub repository has all this documented very well, but here are the highlights:
- Your
srcfolder, containing all your source code. Surprise!
- A directory of
testsfor your code. Always always always always always write tests!
- A
READMEfile of some kind. This is your introduction that summarizes your project, gives a basic overview of how to use it, what the prerequisites are, links to further (more detailed) documentation, and any problems that are known.
- A
docfolder containing detailed documentation about every aspect of your project, including any examples.
- A build script of some sort: a script you can run which automates the majority of the install process.
- Continuous integration (CI). This is bit more advanced, but the idea of continuous integration is to have your code exported out to an auto-compiler every time you update the code, so that you know immediately whether or not it builds successfully.
- A
LICENSEfile. This is often overlooked (like clicking through a EULA), but in the age of open source and Open Science it is absolutely essential to have a license of some sort.
Document, document, document¶
This is drilled into computer science students from day 1, and yet one constant across every software project is that the documentation sucks. To meet basic adequacy standards, documentation should include
- The code itself. This is the radical notion that code should be easy to understand, especially if read by someone who did NOT write it. Save code golf for the competitions!
- Comments in the code. Invariably there will be some logic in the code that doesn't make sense at first glance. This should be explained with clear comments. Not comments like
# loop through the array, but useful comments like# Because the array is already sorted, we only loop through parts of it to search for the value we want.
What does this code do?
def do(a, b):
c = a + " " + b
return c
What if I'd written it this way:
def full_name(first_name, last_name):
name = first_name + " " + last_name
return name
Or even this way:
def full_name(first_name, last_name):
"""
Helper function, to concatenate the first name and the last name
into a single full name.
"""
# Concatenates the two names with a space in between.
name = first_name + " " + last_name
# Return the full name.
return name
Python (and any other programming language) will have a "style" of documenting the code whereby other utilities can take those comments and turn them into documentation.
Java pioneered this approach in the 1990s with "Javadocs" and every language since has emulated that in some way.
Python's version is "Sphinx" http://www.sphinx-doc.org/en/master/

- README. This is usually the user's first introduction to a new software package (GitHub makes this file the default landing page of a repository!). It should clearly state the purpose of the software, how to install and run it, and some basic usage examples before pointing the user to where they can go to learn more.
Examples and tests¶
Right after the README, this is the next place the user will go: examples for how to run the code. Is it command line? What options are there? What kind of data can I run through it? What should I expect for output? These sorts of questions can be addressed with
- Quickstart. This is file that is dedicated to getting the user up and running with the software as quickly as possible with an example. The goal is to put the user in the driver's seat as quickly as possible, and let them ask questions later.
- Examples. This usually takes the form of a bash script, written to invoke the code and have it run on a sample dataset that's provided. Alternatively, you can give a step-by-step guide of what commands to run, what inputs to give, and what the expected output should be.
- Unit tests. You'll learn more about this as you take higher-level software engineering courses, but the main idea is you want to write code that tests your main code. The user can also go here to get a feel for how the code should be used.
- Containers. This is a very advanced topic that we won't cover in this course, but the "wave of the future" is to build your software inside containers (e.g. Docker). Think of containers as super-lightweight virtual machines that anyone can build from a simple script. Using containers, you can package your software in such a way that you don't have to worry about operating systems or clashing software dependencies.
Scientific Notebooks¶
If you're an R or Python user, you've probably heard of "scientific notebooks".
They're interactive, usually web-based applications that blur the lines between
- written documents
- code modules
- figures and tables
by interleaving all three within the same medium.
**These very slides were built entirely in Jupyter, a multi-language scientific notebook platform!
While these slides were built in Jupyter, they were exported into HTML and PDF, so the code isn't technically live anymore.
However, through platforms like mybinder, you can turn any GitHub repository into a collection of active notebooks! You can do this yourself with these slides--all you need is a web browser!

Containers¶
WARNING: This is super-advanced.
Has anyone ever
- tried to run code that is 6+ months old?
- tried to install an old(er) video game on a new computer?
- tried to install a new app on an old smartphone OS?
Did it run?
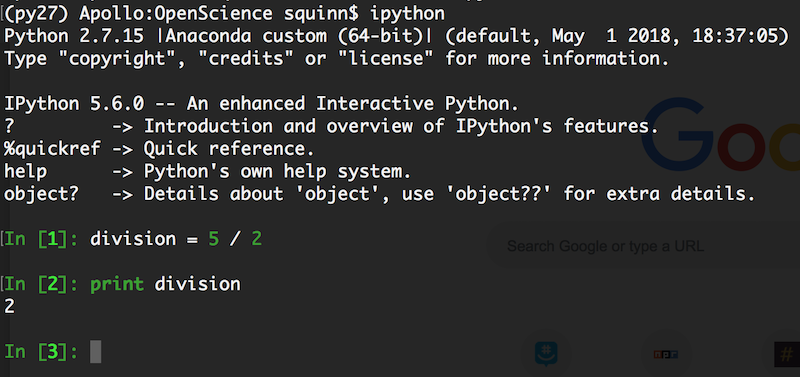
Here's a great example: Python 2 versus Python 3.
Python 2 was the de facto Python version for over a decade (it's kind of the Windows XP of Python). When Python 3 was introduced in the late 2000s, nobody upgraded.
What should this code output?
division = 5 / 2
print(division)
2.5
This notebook is running Python 3. Let's run it in Python 2, shall we?
Containers, such as Docker, allow you to create bash-script-like recipes that build the precise environment you want.

This has huge implications in reproducibility: you can provide others with an exact set of instructions--even a pre-built image!--of the environment in which you conducted your work.
Python Package Index (pypi) is your friend¶

PyPI is the Python Package Index, and is where the vast majority of external Python packages are hosted (over 85,000 as of this lecture!). In addition to publishing your code on GitHub, you can also package your code to be automatically installed via a package manager like pip or easy_install.
Provided you're already adhering to the anatomy of an open source project, there are only a few other steps needed to get your code ready for publishing on PyPI: in particular, writing the setup.py script that will tell the package managers of people who want to install your package exactly how to install your package.
Then you just upload the code to PyPI! http://peterdowns.com/posts/first-time-with-pypi.html
Pre-registration¶
Pre-registration means publishing your methods publicly BEFORE any work has been done.
Why is this important?
Sites like Protocols.io https://www.protocols.io/ and OSF https://osf.io/prereg/ have pre-registration modules, sometimes linked with certain journals who agree to publish the results of the methods, regardless of whether they're positive or negative.


This makes sense for wet lab / life sciences or even theory-driven statistics, but what about empirical or computational studies like applied machine learning?
Turns out, there are a number of pre-registration sites for those too, including OpenML.

Licensing¶
Yes, this is super boring, I agree. Fortunately, the wonderful folks at GitHub have created an awesome resource for you to use.
Choose a License: It provides a handy choose-your-adventure flowchart for picking the perfect open software license for your project. I highly recommend it.
However, if you're just looking for a tl;dr version and want the basic lay of the land, GitHub's default licenses for new projects include
- MIT License (pretty much the "no warranty whatsoever, use at your own risk" license)
- Apache 2.0 License (almost exactly like the MIT, but allows for patents and trademarks)
- GPLv3 License (requires anyone who modifies your code to publish those modifications under GPLv3 too)
Final Thoughts¶
In my view, Open Science and its practices will only become more important.
- Replicating previous results
- Providing hands-on tools to the upcoming generation of scientists
Six major areas of Open Science
- These have to be addressed from the beginning, not tacked on after the fact
Already lots of great open source tools and repositories to encourage Open Science practices
- data repositories
- tools for method replication and testing
- standards for code structure, results presentation, and method preregistration
Administrivia¶
- How is Assignment 5 going? Due TONIGHT at 11:59pm!
- This was the last lecture of the course! Congratulations!
- Final project talks will be ext week on Tuesday and Thursday!
Additional Resources¶
- GitHub template for open source Python projects https://github.com/uwescience/shablona
- Mozilla Open Science Lab https://science.mozilla.org/
- Open Science Framework (OSF) for all phases of the scientific pipeline https://osf.io/
- The Center for Open Science (COS), which supports OSF https://cos.io/
- Docker https://www.docker.com/