Lecture 1: Welcome and Introduction¶
CBIO (CSCI) 4835/6835: Introduction to Computational Biology
Overview and Objectives¶
In this lecture, we'll define and discuss the field of "computational biology" and motivate programming in a wet lab setting. We'll also go over the logistics of CBIO 4835/6835 and how the course will proceed over the semester. By the end of this lecture, you should be able to
- Define "computational biology" and the importance of having coding skills
- Define the Python programming language
- Take the course pre-test (Google doc, linked at the end)
- Log into the course Slack channel and JupyterHub
What is "Computational Biology"?¶
Is it "biology, but with computers"?
What does that even mean, anyway? Is using Excel considered "computational"?
What differentiates biology from computational biology?
What about quantitative biology? Where does that fit in?
Is that a subset of computational biology, or is computational biology a subset of it? Or do they just share some overlap?
And isn't all this just "bioinformatics"?

For the purposes of this course, I'm less concerned with properly categorizing computational biology vs bioinformatics, and more interested in
- Giving everyone the opportunity to gain experience in programming
- Teaching Python
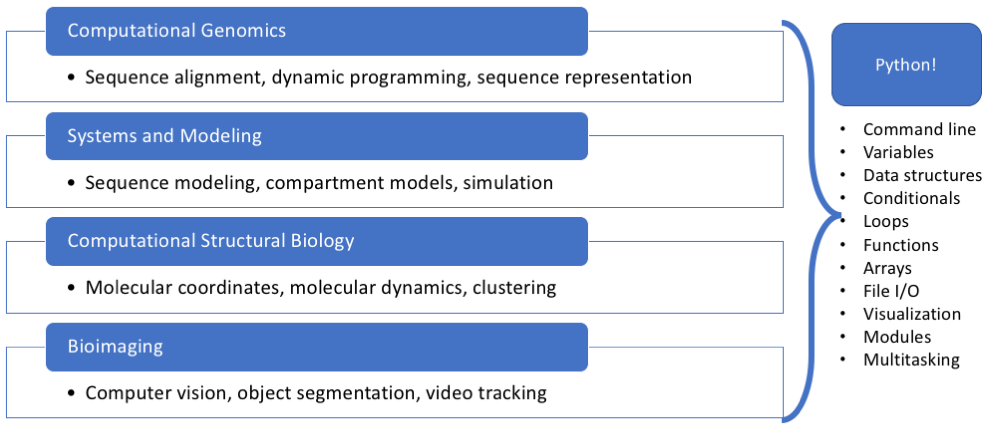
- Surveying computational methods in a biological context
- Improving everyone's skills to be more productive and successful researchers
What this course is¶
Officially: Introduction to Computational Biology.
Officially, but more colloquially: programming for biologists.
What this course is not¶
Biology for programmers.
What this means¶
I assume little or NO knowledge of
- programming or command-line scripting
- probability and statistics
- computational biology
I assume substantive knowledge of
- molecular and cellular biology
- chemistry and biochemistry
If you're here to learn new computational approaches and incorporate them into your current lab work, welcome!¶
If you're here to learn biology to apply your existing computational knowledge, you may want to look elsewhere¶
Why Python?¶


Python¶
The language is designed from the ground-up to be easy to use.
It's a full-featured (like C++ or Java), powerful language that can be used in a lot of different contexts.
Most importantly, it's free: a completely open-source platform that costs nothing.
Python is also an extremely popular language.

Of course, while that may turn off the programming language hipsters, it does mean that the language and its surrounding ecosystem has lots of momentum. This comes in handy when we want to explore specific problems!
Between 2017 and 2018, Python actually attained the status of Most Popular Programming Language!
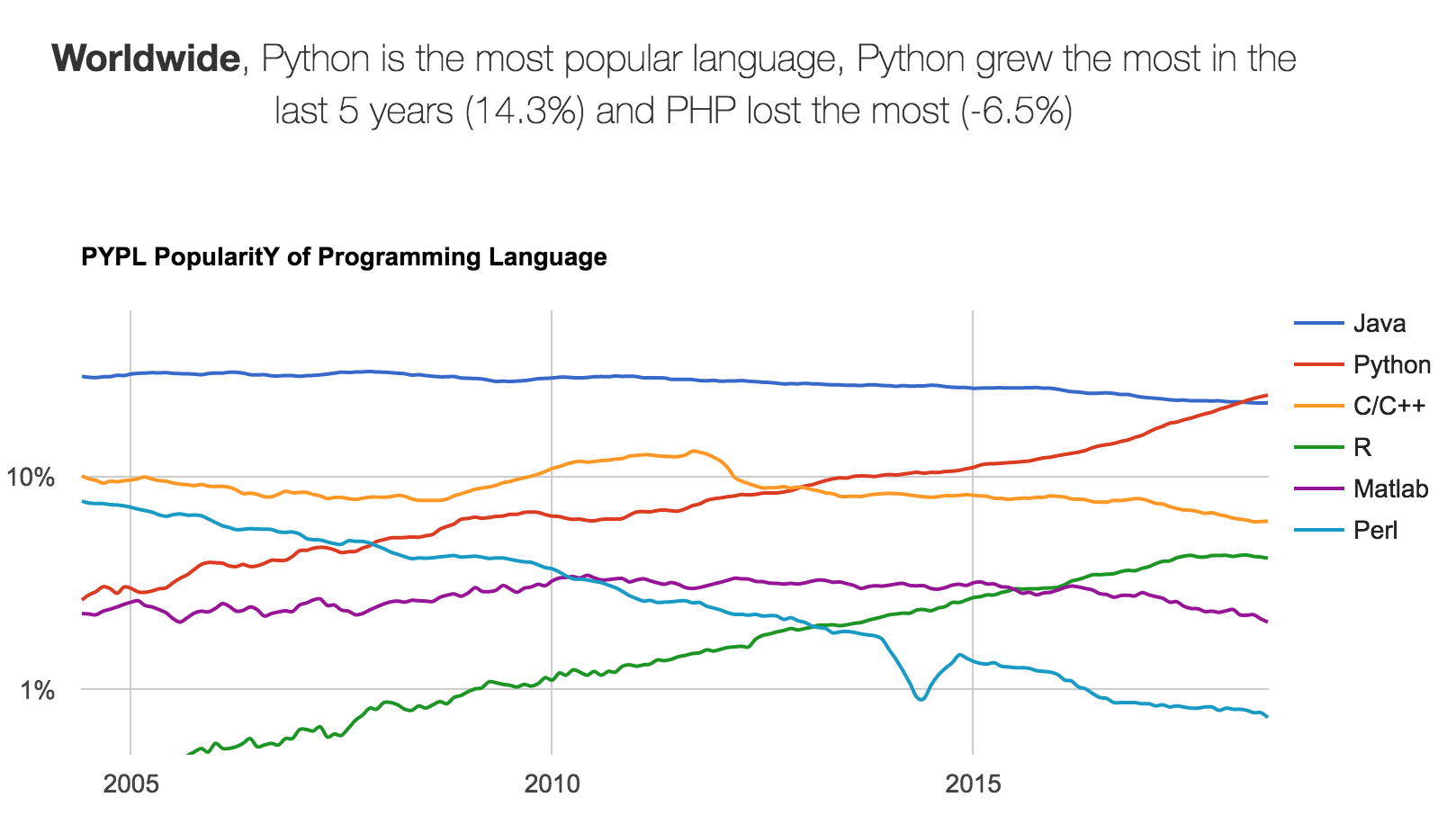
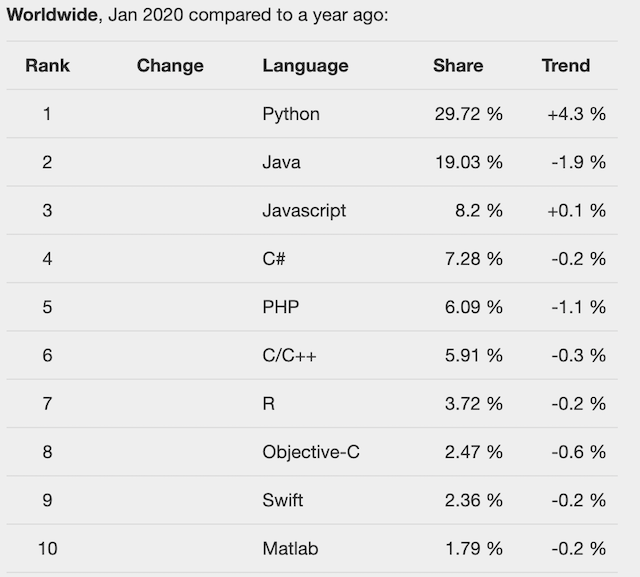
See more programming language statistics here: http://pypl.github.io/PYPL.html
... but why Python?¶
The ecosystem!
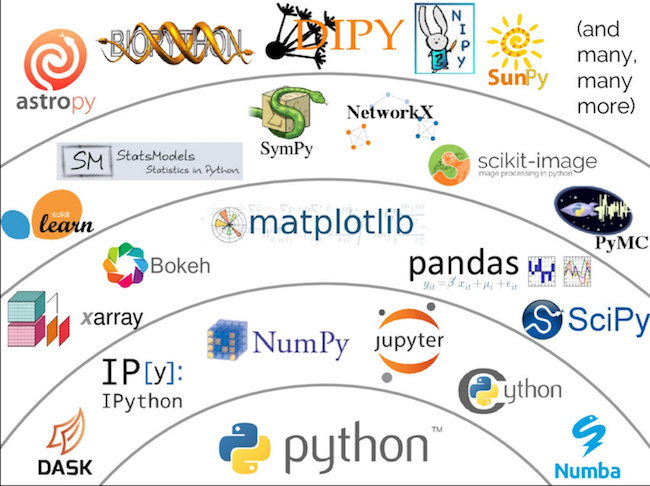
Python is cool, Computational Biology is a thing...so what?¶
Let's look at the current wet lab experimental pipeline.

Seems good enough. Now, let's add a wrinkle.

Oh, goodie. Back to the lab for another round of sleepless nights.
You can imagine how this can go on and on. Wouldn't it be nice to collect data once and, I don't know, automate the analysis?
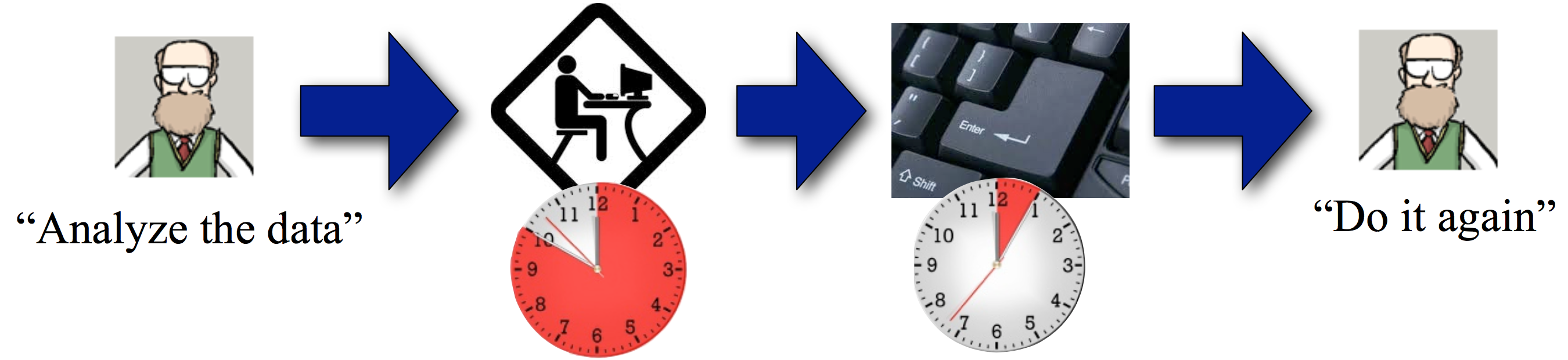
It's that step at the end that's key.
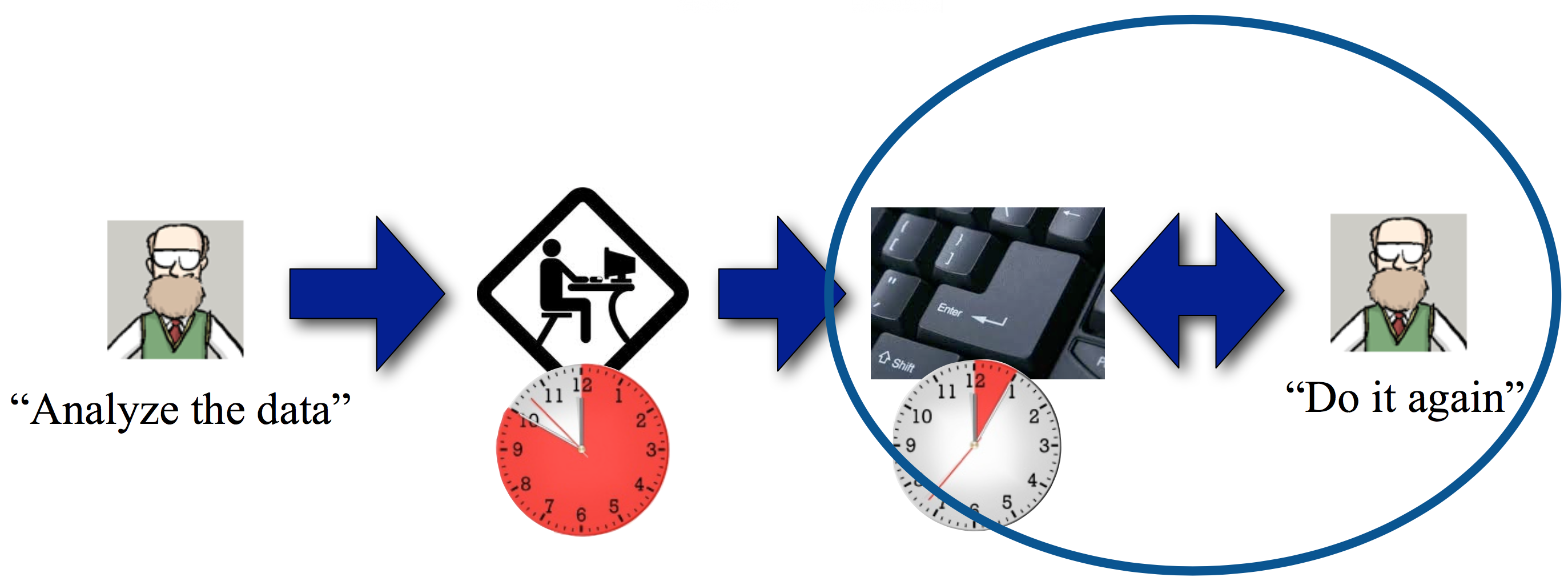
In some sense, it's the goal of computational biology to become biology--that is, "computational biology" will be a redundant way of referring to the overall field.
If anyone is familiar with the buzzphrase of the 2010s "data science", you can think of computational biology as data science in biology.
In this course, we'll combine programming in Python with statistics to automate analyses of biological data.
Course Logistics¶
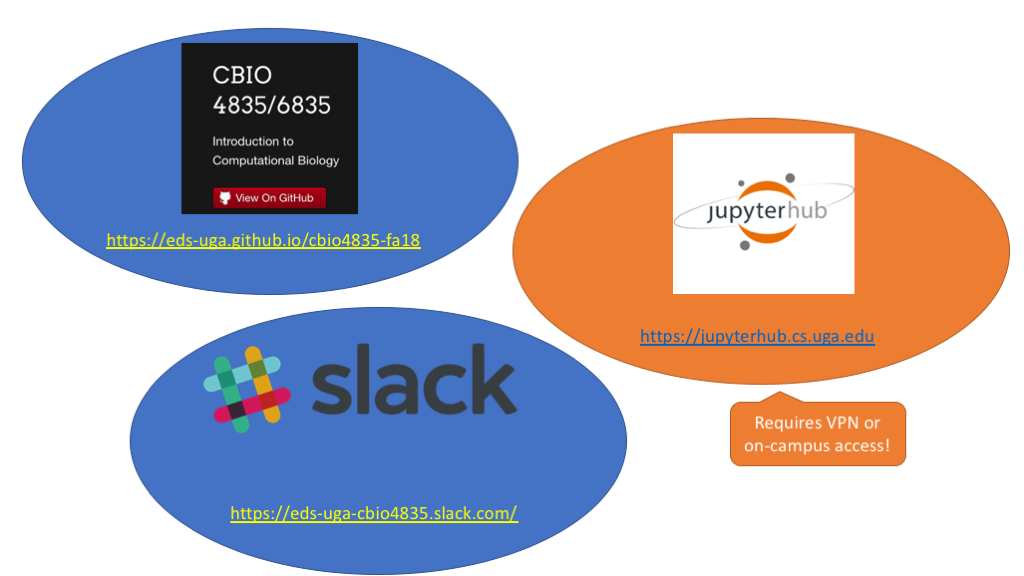
All lecture materials will be posted on the course website:
You do NOT have to install Python on your own computer; the only requirement is that you have something with a modern web browser and an internet connection.
However, installing Python on your own machine does give you more tinkering elbow room, though (instructions to this effect to follow in a future lecture).
Lectures¶
Attendance is NOT mandatory. You're all adults.
That said, my least favorite question is from a student I've never seen in lecture coming to office hours for the first time the day before the midterm, asking me to summarize the semester for them.
Make yourself a regular in lecture, a regular on the Slack chat (asking AND answering questions), and letting me know when you need to miss lecture (we've all got things to do; you don't have to ask permission, just let me know when you won't be there), and you'll be fine.
Grading¶
The breakdown is summarized in the course syllabus (linked on the website, also found here: https://eds-uga.github.io/cbio-x835-sp20/syllabus.pdf).
Assignments¶
There will be 6 assignments. They'll be released on Thursdays and due by 11:59pm two weeks later.
They will all be released over JupyterHub in the form of Jupyter notebooks.
We'll go more over this format in a future lecture, but suffice to say you'll do the assignments in the entirety through a web browser. Thus, you won't need to install Python on your own machine unless you want to.
Projects¶
Final projects will consist of three main components:
- A brief proposal, outlining the project you want to run, the dataset you'll use, and the computational experiments you'll perform
- A 15-minute presentation at the end of the semester, outlining your major results
- A full conference/journal-ready paper, detailing the problem you worked on, the methods you used, and the results you obtained
More details will be released later in the semester. Be thinking about what you might want to do!
JupyterHub¶
Assignments will be released, completed, and submitted by JupyterHub. Everyone should have their own login (if you don't or it isn't working, let me know!).
The assignments will be in Jupyter notebook format. Jupyter notebooks will come with their own autograders, so you can run those tests on your completed sections before submitting. Occasionally there are errors in the autograders, so if you suspect your code is correct even though the autograder is throwing errors, post about it in Slack!
You'll subsequently submit completed Jupyter notebooks through JupyterHub as well. This final step is critical; you have to click "Submit" for me to see your assignment and give it a grade! In the past this has only rarely been a problem, but nonetheless something to keep in mind.
JupyterHub is more than just a conduit for homework assignments. You can also create your own Jupyter notebooks and experiment with Python! This is a great alternative to installing Python on your own computer. I highly encourage you to do this!
Slack¶
Slack is the primary way we'll keep in touch over the semester.
- I'll post announcements in the
#announcementschannel (please keep it clear). - If you have any questions related to course concepts or homework problems, please post in
#questions. - If you're unable to log in, or are getting strange errors you can't diagnose, let me know in
#techprobs. - If you found a certain topic in class interesting and want to discuss it further, or found a cool article related to computational biology, or something else entirely, feel free to strike up a discussion in the
#lounge.
You can also DM me or your classmates directly (this is WAY better than email for me!).
Office Hours¶
My office is located in Boyd GSRC, room 638A.
Office hours are current TBD, but they'll be in my office (surprisingly).
You are also welcome to set up a separate appointment with me! DM me over Slack or shoot me an email to set up a time to meet.
Academic Honesty¶
Do your own work. Programming and coding is a lot like writing--everyone has their own style, so it's very easy to spot copied work.
That said, please do discuss concepts and problem solving strategies! Either during in-person group meetings or over group chats on Slack.
One reason I like Slack is because I can jump in when I see a question, but if I'm otherwise occupied someone else might answer it first.
Pre-test¶
This is an ungraded survey that will help me to assess everyone's background and properly calibrate the course. It's ungraded, but it is required that everyone finish it by Thursday's lecture. (Let's call it: "Assignment 0")
https://docs.google.com/forms/d/1ka9yH5G3bOCfdJUTaeZXV2BdtvqqsiPaxnvKI2f4YK4/
Administrivia¶
- "Assignment 0" due Thursday
- Finish the pre-test
- Read the syllabus
- Make sure you can log into JupyterHub
- Make sure you can access Slack
Questions?¶
Questions? Check the course website: https://eds-uga.github.io/cbio-x835-sp20
Still more questions? Ask on the Slack chat: https://eds-uga-cbio4835.slack.com/
Still more questions? Email me: squinn@cs.uga.edu (include CBIO 4835 or CBIO 6835 in the Subject line so it hits my filters)
Still more questions? Erm...
